Variant Discovery
In order to call a variant, it must have been 'discovered' and included in the set of candidate variants. Therefore, the final called variants is a subset of candidate variants. To maximise sensitivity, Octopus uses a hybrid approach to variant discovery by combining candidates discovered from multiple strategies.
Pileups#
The pileup candidate generator discovers variants directly from input alignments (i.e. the mapper CIGAR entries). Mismatch and indel positions are aggregated for filtered input reads, and candidates are accepted based on several criteria.
The pileup candidate generator can be disabled with the --disable-pileup-candidate-generator option.
Repeat expander#
Similar to the pileup candidate generator, the repeat expander candidate generator examines input alignments, but looks for specific patterns of SNV mismatches in compound tandem repeat regions (tandem repeats composed of two or more motifs) that are indicative of misaligned balanced indels. An example is shown below:
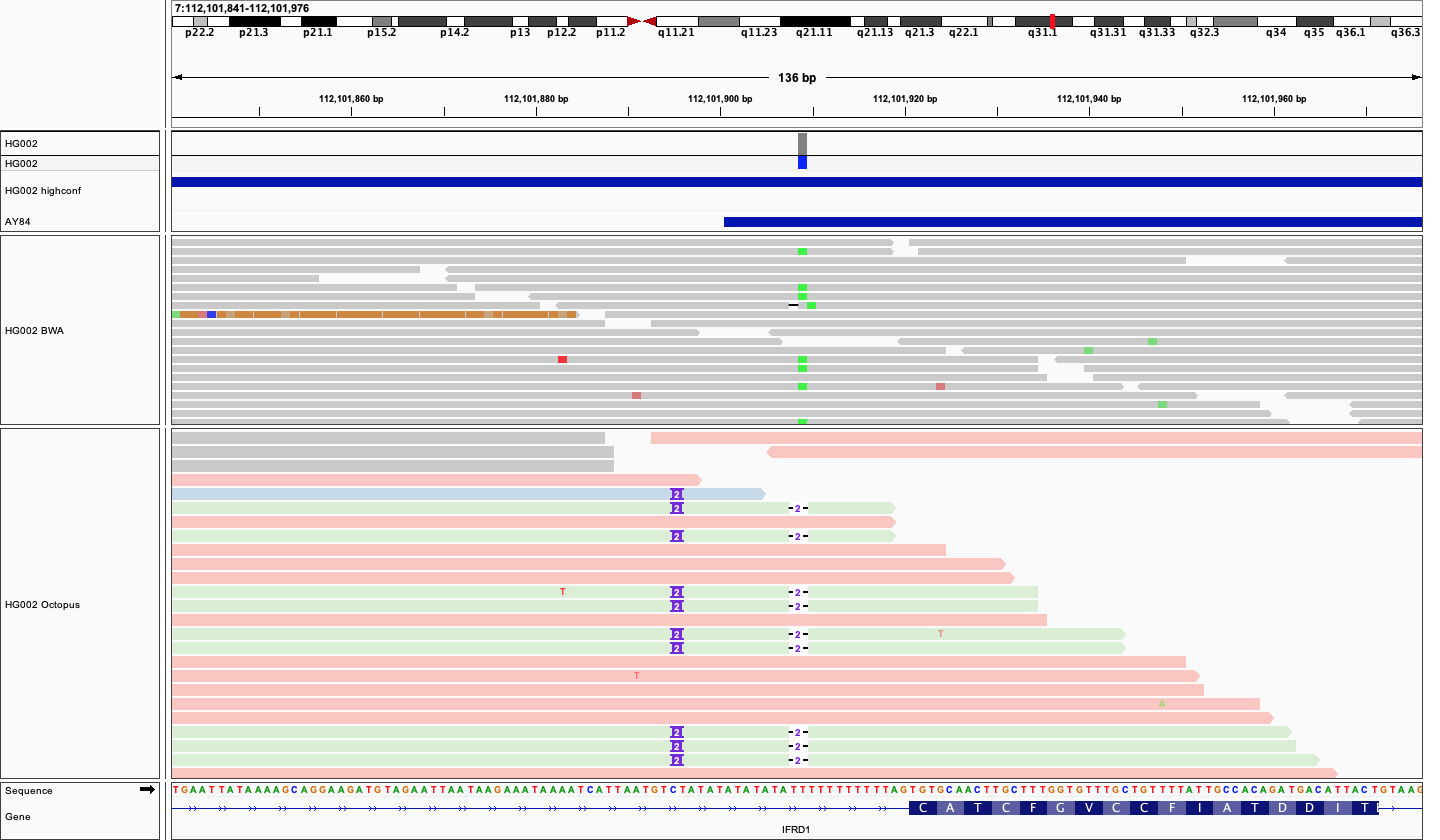
The repeat expander candidate generator can be disabled with the --disable-repeat-candidate-generator option.
Local reassembly#
When input reads are misaligned, the pileup generator may propose false candidates. In such cases, correct variants can still be discovered by assembling reads with a de-Bruijn graph. Both the reference sequence and reads are threaded into the graph and any 'bubbles' (divergences from the reference path) are explored (up to --max-bubbles) and scored by counting k-mer observations. To help remove spurious bubbles and increase sensitivity, mismatch bases with low base quality scores (--assembler-mask-base-quality) are converted to reference before graph threading into the assembly graph. Bubbles with a high score (minimum --min-bubble-score) are extracted from the graph, before the alignment to the reference portion of the bubble with Smith Waterman to find candidate variants.
To simplify construction and evaluation, graphs that contain cycles in the reference sequence are not not allowed. This restriction necessitates the use of relatively small reference subsequences (up to --max-assembly-region-size), though cycles can also be avoided with large k-mer sizes. As such, for a given assembly region, assembly is attempted with the multiple k-mer sizes specified with --kmer-sizes. If none of these k-mer sizes result in an acylic graph, then assembly is attempted with up to --num-fallback-kmers. By default, graphs containing non-reference cycles are also not allowed as this often results in spurious candidates. However, this restriction can be disabled with the --allow-cycles command.
De novo assembly is significantly more complex and time consuming than evaluating pileups, therefore by default assembly is only attempted using reads mapped to small genomic regions (hence local assembly). In addition, only regions considered to have a high likelihood of containing variation are assembled (unless the --assemble-all command is specified).
tip
The maximum assembly region size (--max-assembly-region-size) implicitly limits the maximum discoverable deletion and complex substitution size. However, using larger region sizes increases the chance of a reference cycle in the assembly graph, necessitating the use of larger k-mer sizes, which can in turn hurt sensitivity.
The assembly candidate generator can be disabled with the --disable-assembly-candidate-generator option.
Input VCF#
Previously called variants in VCF can be included as candidates with the --source-candidates and --source-candidates-file options. By default, only records with FILTER set to PASS or . are included. To use all variants the --use-filtered-source-candidates should be specified.
important
Supplying input candidates does not guarantee that they will be reported in the final VCF - they are treated just like candidates discovered from the other generators. Octopus does not currently provide complete 'regenotyping' functionality. However, using the --disable-denovo-variant-discovery command along with the --source-candidates is a reasonable approximation - the output VCF will be a subset of the input.

 Octopus
Octopus